Department of Peptidomics
Member
You can scroll horizontally.
| Chief (Principal Investigator) | ||
| Research Scientist | Masao Nakamura, Ph.D. | Researchmap |
| Research Assistant | ||
| Visiting Scientist | Kazuki Sasaki, M.D., Ph.D. | Tochigi Cancer Center Research Institute Researchmap |
| Research Student | Momoko Nakamura | Hoshi University |
| Research Student | Tomonori Hariya | Hoshi University |
| Research Student | Arisa Yamaguchi | Hoshi University |
*Please add @po.kyoundo.jp after e-mail address
Overview
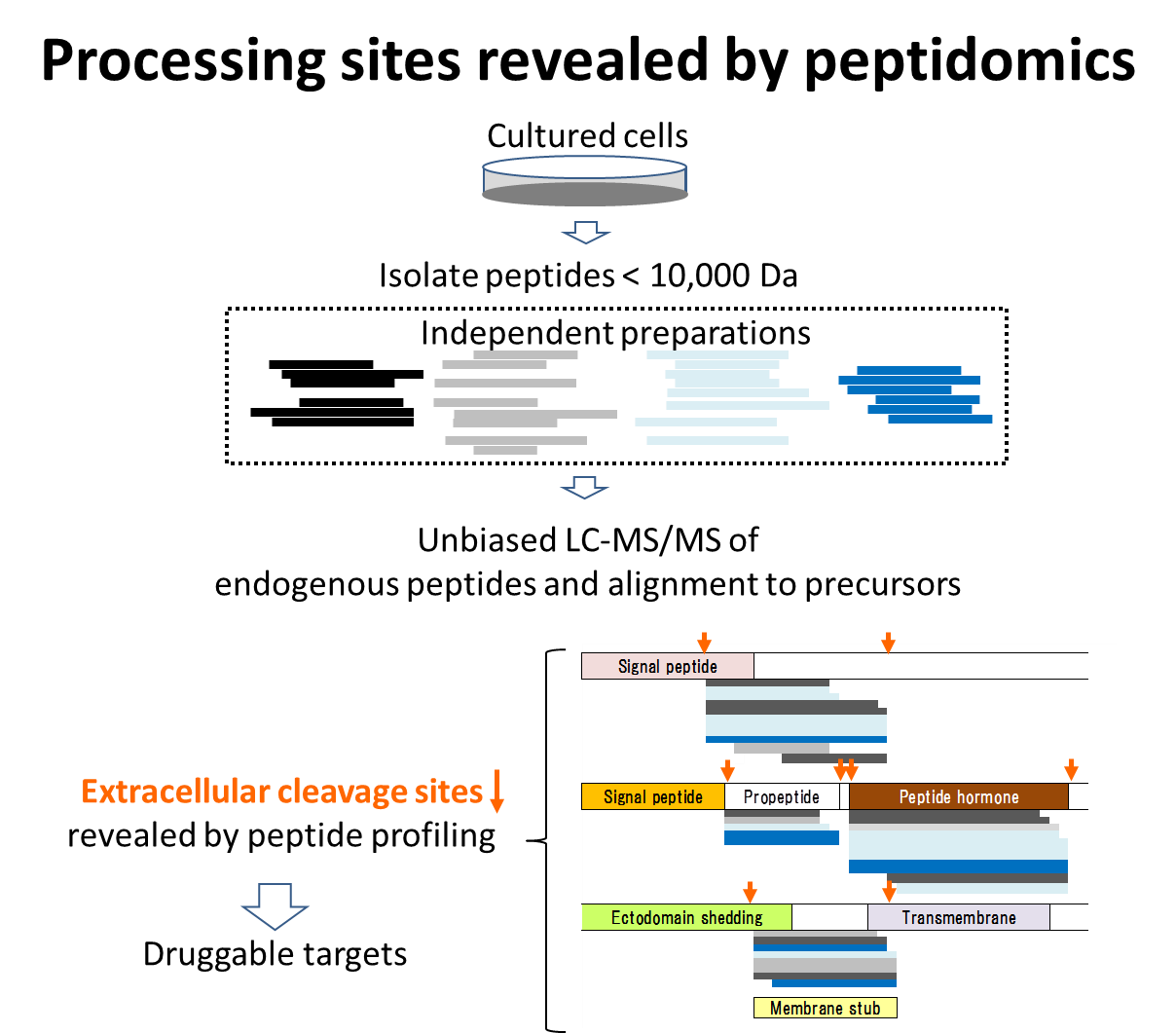
Peptidomics is a comprehensive study of naturally occurring peptides smaller than 10,000 Da. Apart from peptide hormones and anti-microbial peptides, they are in a relatively uncharted territory of the proteome. We have developed a platform for profiling endogenous peptides using mass spectrometry. Our profiling study helps to elucidate proteolytic processing sites of secretory proteins and transmembrane proteins, marking the locations where peptide hormones are cleaved off, or soluble receptors are created to be a ligand sink, to name a few. Given that the processing is not just regulated by particular sequences but by post-translational modifications, like glycosylation, neighboring the cleavage sites, the whole picture is so complicated that in silico prediction falls well short of our expectation. This situation makes peptidomics the method of choice for delineating processing sites in an unbiased manner. Our earlier study to define procesussing sites of secretory proteins led to the identification of biologically active peptides functioning in the nervous and endocrine systems. We are currently looking at transmembrane proteins that undergo extracellular processing with the aim of identifying potential targets applicable to the therapeutics and diagnostics of pancreatic ductal adenocarcinoma. Unlike other omics, peptidomics has lagged behind and remained underrepresented, but this study is part of a larger pattern we are going to see over the years to come.
Projects
Our efforts are directed towards the identification of transmembrane proteins that could serve as a potential target for treating pancreatic ductal adenocarcinoma. Some transmembrane proteins undergo ectodomain shedding, leaving behind membrane stubs on the cell surface. These membrane-associated segments can be a potential target of antigen-dependent cellular cytotoxicity. We have demonstrated that peptidomics-based endogenous peptide profiling helps to identify proteolytic processing sites of secretory or transmembrane proteins. This is accomplished by mass spectrometric sequencing of peptides larger than 3,000 Da, which are beyond the reach of current bottom-up proteomics.
Publications
Last 5 years
- Hayashi M, Noguchi R, Abe M, Osaki J, Adachi Y, Iwata S, Sasaki K, Kondo T, Yoshimatsu Y: Gastric biopsy-derived cell line and its utility in assessing tumor cell drug sensitivity. Biomed Res 46: 27-35, 2025.
- Noguchi R, Osaki J, Ono T, Adachi Y, Iwata S, Yoshimatsu Y, Sasaki K, Kawai A, Kondo T: Pharmacoproteogenomic approach identifies on-target kinase inhibitors for cancer drug repositioning. In Vitro Cell Dev Biol Anim 60: 1200-1214, 2024.
- Nakamura M: Development of therapeutic antibody enhancers through glycan cleavage in refractory cancers. Impact 2: 22-23, 2024.
- Zhang W#, Miura A#, Abu Saleh MM, Shimizu K, Mita Y, Tanida R, Hirako S, Shioda S, Gmyr V, Kerr-Conte J, Pattou F, Jin C, Kanai Y, Sasaki K, Minamino N, Sakoda H, Nakazato M: The NERP-4-SNAT2 axis regulates pancreatic β-cell maintenance and function. Nature Commun 14: 8158, 2023. #Contributed equally.
- Nakamura M, Shiga A, Iimori A, Matsuzaki T: Efficient endocytosis of the human lactoferrin N-lobe enhances its antiproliferative activity against human cancer cells. Biol Pharm Bull. 46: 1004-1009, 2023.
- Paudel D, Kuramitsu Y, Uehara O, Morikawa T, Yoshida K, Giri S, Islam S, Kitagawa T, Kondo T, Sasaki K, Matsuoka H, Miura H, Abiko Y: Proteomic and microbiota analyses of the oral cavity during psychological stress. PLoS ONE 17: e0268155, 2022.
- Nakamura M, Sato A: Glycan-binding properties of basic whey protein lactoferrin and its application in nerve regenerative medicine. Trends Glycosci. Glycotechnol. 34: E19-E23, 2022.
- Miyamoto S, Nagano Y, Miyazaki M, Nagamura Y, Sasaki K, Kawamura T, Yanagihara K, Imai T, Ohki R, Yashiro M, Tanaka M, Sakai R, Yamaguchi H: Integrin α5 mediates cancer cell-fibroblast adhesion and peritoneal dissemination of diffuse-type gastric carcinoma. Cancer Lett. 526: 335-345, 2022.
- Nakamura M: Development of a highly functionalized lactoferrin that controls nerve function: Fusion of glycan-binding ability and neuroprotective function. Glycoforum 24: A8, 2021.
- Nakamura M, Matsuzaki T, Iimori A and Sato A: Harnessing the chondroitin sulfate-binding characteristics of human lactoferrin to neutralize neurite outgrowth inhibition. Biochem. Biophys. Res. Commun. 534: 1076-1082, 2021.
- Nagamura Y, Miyazaki M, Nagano Y, Yuki M, Fukami K, Yanagihara K, Sasaki K, Sakai R, and Yamaguchi H: PLEKHA5 regulates the survival and peritoneal dissemination of diffuse-type gastric carcinoma cells with Met gene. Oncogenesis 10: 25, 2021.
- Miyazaki S, Tashiro F, Tsuchiya T, Sasaki K and Miyazaki JI: Establishment of a long-term stable β-cell line and its application to analyze the effect of Gcg expression on insulin secretion. Sci. Rep. 11: 477, 2021.
- Tsuchiya T, Nakayama A, Kawamura T and Sasaki K: Capillary electrophoresis electrospray ionization-mass spectrometry for peptidomics-based processing site determination. Biochem. Biophys. Res. Commun. 533: 872-878, 2020.
- Ueda K, Shimizu M, Ohashi A, Murata D, Suzuki T, Kobayashi N, Baba J, Takeuchi T, Shiga Y, Nakamura M, Kagaya S and Sato A: Albumin fusion at the N-terminus or C-terminus of human lactoferrin leads to improved pharmacokinetics and anti-proliferative effects on cancer cell lines. Eur. J. Pharm. Sci. 155: 10551, 2020.
Before 2020 (Selected)
- Matsuzaki T, Nakamura M, Nogita T and Sato A: Cellular uptake and release of intact lactoferrin and its derivatives in an intestinal enterocyte model of Caco-2 cells. Biol. Pharm. Bull. 42: 989-995, 2019.
- Sasaki K, Tsuchiya T, and Osaki T: Isolation of Endogenous Peptides from Cultured Cell Conditioned Media for Mass Spectrometry. Methods Mol. Biol. 1719: 51-58, 2018.
- Tsuchiya T, Osaki T, Minamino N, and Sasaki K: Peptidomics for studying limited proteolysis. J. Proteome Res. 14: 4921-4931, 2015.
- Nonaka M, Kim R, Fukushima H, Sasaki K, Suzuki K, Okamura M, Ishii Y, Kawashima T, Kamijo S, Takemoto-Kimura S, Okuno H, Kida S, and Bito H: Region-Specific Activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron 84: 92-106, 2014.
- Sasaki K, Osaki T and Minamino N: Large-scale identification of endogenous secretory peptides using electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 12: 700-709, 2013.
- Nakamura M, Uehara Y, Asada M, Suzuki M and Imamura T: Sulfated glycosaminoglycan-assisted receptor specificity of human fibroblast growth factor (FGF) 19 signaling in a mouse system is different from that in a human system. J. Biomol. Screen. 18: 321-330, 2013.
- Shimamura S, Sasaki K and Tanaka M: The Src substrate SKAP2 regulates actin assembly by interacting with WAVE2 and cortactin proteins. J. Biol. Chem. 288: 1171-1183, 2013.
- Nagai N, Habuchi H, Sugaya N, Nakamura M, Imamura T, Watanabe H and Kimata K: Involvement of heparan sulfate 6-O-sulfation in the regulation of energy metabolism and the alteration of thyroid hormone levels in male mice. Glycobiology 23: 980-992, 2013.
- Osaki T, Sasaki K and Minamino N: Peptidomics-based discovery of an antimicrobial peptide derived from insulin-like growth factor-binding protein 5. J. Proteome Res. 10: 1870-1880, 2011.
- Nakamura M, Uehara Y, Asada M, Honda E, Nagai N, Kimata K, Suzuki M and Imamura T: Sulfated glycosaminoglycans are required for specific and sensitive fibroblast growth factor (FGF) 19 signaling via FGF receptor 4 and betaKlotho. J. Biol. Chem. 286: 26418-26423, 2011.
- Sasaki K, Takahashi N, Satoh M, Yamasaki M and Minamino N: A peptidomics strategy for discovering endogenous bioactive peptides. J. Proteome Res. 9: 5047-5052, 2010.
- Sasaki K, Satomi Y, Takao T and Minamino N: Snapshot peptidomics of the regulated secretory pathway. Mol. Cell. Proteomics 8: 1638-1647, 2009.
- Yamaguchi H, Sasaki K, Satomi Y, Shimbara T, Kageyama H, Mondal MS, Toshinai K, Date Y, González LJ, Shioda S, Takao T, Nakazato M and Minamino N: Peptidomic identification and biological validation of neuroendocrine regulatory peptide-1 and -2. J. Biol. Chem. 282: 26354-26350, 2007.
- Sasaki K, Sato K, Akiyama Y, Yanagihara K, Oka M and Yamaguchi K: Peptidomics-based approach reveals the secretion of the 29-residue COOH-terminal fragment of the putative tumor suppressor protein DMBT1 from pancreatic adenocarcinoma cell lines. Cancer Res. 62: 4894-4898, 2002.